This paper proposed a theoretically plausible dropout method for RNNs. When applied to language modeling, the new technique outperforms existing approaches.
Introduction
Dropout is a popular regularization technique which prevents deep networks from overfitting. But empirical results have shown the negative effects of applying dropout in RNNs' recurrent connections. Only dropout in non-recurrent connections has been shown to improve performance. So, people often believe that noise added to recurrent layers will be amplified across long sequences which drown the signal.
The authors proposed a new dropout variant, which repeats the same dropout mask at each time step, at each layer, for both inputs and outputs, as shown in Figure 1:
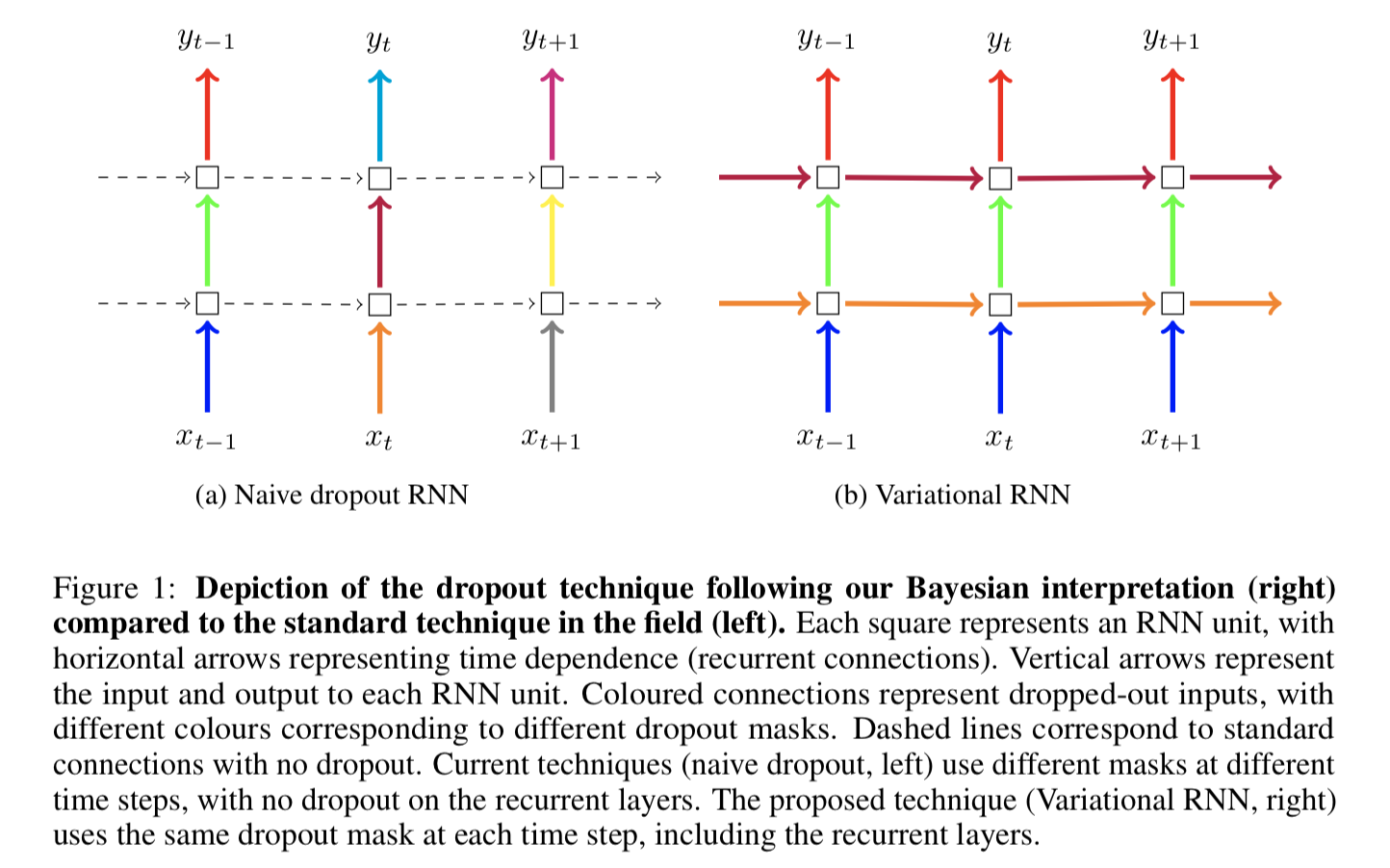
For figure to the right, blue connects share the same dropout mask, then green ones, then red ones. Those dropout masks are in fact dropping recurrent units (words) out. Differently, the purple and yellow masks are applied to hidden states across time steps.
Background
This variant is variational inference based, what does this mean?
Bayesian Neural Networks
In Bayesian regression, given data inputs \(\mathbf{X} = \{ \mathbf{x_1}, \cdots, \mathbf{x}_N \}\) and outputs \(\mathbf{Y} = \{\mathbf{y}_1, \cdots, \mathbf{y}_N\}\), we want to find the parameters \(\omega\) which are likey to have generated that data. Following the Bayesian approach, we put some prior distributions \(p(\omega)\) over the parameter space.
The likelihood distribution \(p(\mathbf{y} | \mathbf{x}, \omega)\) for classification is:
\[p(y = d|{\mathbf{x}},\omega ) = {\exp (f_d^\omega ({\mathbf{x}}))/\sum\limits_{d'} {\exp } (f_{d'}^\omega ({\mathbf{x}}))}\]
The authors wrote a different equation:
\[p(y = d|{\mathbf{x}},\omega ) = {\text{Categorical}}\left( {\exp (f_d^\omega ({\mathbf{x}}))/\sum\limits_{d'} {\exp } (f_{d'}^\omega ({\mathbf{x}}))} \right)\]
I don't know what does that mean.
Anyway, the posterior distribution over parameter space \(p(\omega \vert \mathbf{X}, \mathbf{Y})\) is then used to predict a new input point by integrating
\[
p({{\mathbf{y}}^*}|{{\mathbf{x}}^*},{\mathbf{X}},{\mathbf{Y}}) = \int p ({{\mathbf{y}}^*}|{{\mathbf{x}}^*},\omega )p(\omega |{\mathbf{X}},{\mathbf{Y}}){\text{d}}\omega
\]
If you place a prior distribution over a neural network's weights, then that NN is called a Bayesian NN. The common approach is to place gaussian prior \( p({{\mathbf{W}}_i}) = \mathbb{N}({\mathbf{0}},{\text{ }}{\mathbf{I}})\) over weights, to assume point estimate for the bias.
Approximate Variational Inference in Bayesian NN
What's Variational Inference
Given dataset, the posterior over parameters \(p(\omega\vert \mathbf{X}, \mathbf{Y})=\frac{p(\mathbf{X},\mathbf{Y}, \omega )}{p(\mathbf{X},\mathbf{Y})}=\frac{p(\mathbf{X},\mathbf{Y}\vert \omega)p(\omega )}{\int p(\mathbf{X},\mathbf{Y}\vert \omega)p(\omega ) \text{d}\omega }\) is not tractable. The denominator integrates over entire parameter space. To approximate, we find a tractable \(q(\omega\vert \mathbf{X}, \mathbf{Y})\), and minimize the KL divergence between \(p(\omega\vert \mathbf{X}, \mathbf{Y})\) and \(q(\omega\vert \mathbf{X}, \mathbf{Y})\).
KL divergence
\[\begin{equation} \begin{aligned}
{\mathbf{KL}}(q(\omega )||p(\omega |{\mathbf{X}},{\mathbf{Y}})) &= - \int q (\omega )\log \frac{{p(\omega |{\mathbf{X}},{\mathbf{Y}})}}{{q(\omega )}}d\omega \hfill \\
&\propto - \int q (\omega )\log \frac{{p({\mathbf{Y}}|{\mathbf{X}},\omega )p(\omega )}}{{q(\omega )}}d\omega \hfill \\
&\propto - \int q (\omega )\log p({\mathbf{Y}}|{\mathbf{X}},\omega )d\omega - \int q (\omega )\log \frac{{p(\omega )}}{{q(\omega )}}d\omega \hfill \\
&\propto - \int q (\omega )\log p({\mathbf{Y}}|{\mathbf{X}},\omega )d\omega + {\mathbf{KL}}(q(\omega )||p(\omega )) \hfill \\
&={ - \sum\limits_{i = 1}^N {\int q } (\omega )\log p({{\mathbf{y}}_i}|{{\mathbf{f}}^\omega }({{\mathbf{x}}_i})){\mathbf{d}}\omega + {\mathbf{KL}}(q(\omega )||p(\omega ))}
\end{aligned} \end{equation} \]
Variational Inference in Recurrent Neural Networks
Given input sequence \({\mathbf{x}}{\text{ = [}}{{\mathbf{x}}_1}{\text{, ..., }}{{\mathbf{x}}_{\text{T}}}{\text{]}}\) of length \(T\), GRU is such a repeated application of function \(\mathbf{f}_{\mathbf{h}}\):
\[
{{\mathbf{h}}_t}{\text{ }} = {\text{ }}{{\mathbf{f}}_{\mathbf{h}}}({{\mathbf{x}}_t},{\text{ }}{{\mathbf{h}}_{t - 1}}) = \sigma ({{\mathbf{x}}_t}{\text{ }}{{\mathbf{W}}_{\mathbf{h}}} + {\text{ }}{{\mathbf{h}}_{t - 1}}{\text{ }}{{\mathbf{U}}_{\mathbf{h}}} + {\text{ }}{{\mathbf{b}}_{\mathbf{h}}})
\]
Then the model output can be defined as a affine transform of the last hidden state:
\[
{{\mathbf{f}}_{\mathbf{y}}}({{\mathbf{h}}_T}) = {{\mathbf{h}}_T}{{\mathbf{W}}_{\mathbf{y}}} + {{\mathbf{b}}_{\mathbf{y}}}
\]
To view the GRU as a probabilistic model, we treat \( \omega = \{ {{\mathbf{W}}_{\mathbf{h}}},{{\mathbf{U}}_{\mathbf{h}}},{{\mathbf{b}}_{\mathbf{h}}},{{\mathbf{W}}_{\mathbf{y}}},{{\mathbf{b}}_{\mathbf{y}}}\} \) as random variables. Denote that \({\mathbf{f}}_{\mathbf{y}}^\omega = {{\mathbf{f}}_{\mathbf{y}}}\) to clarify the dependency on \(\omega \).
Use variational inference with distribution \(q(\omega)\) to approximate posterior over \(\omega\). Evaluate each sum term in eq. (2):
\[\begin{equation} \begin{aligned}
\int q (\omega )\log p({\mathbf{y}}|{\mathbf{f}}_{\mathbf{y}}^\omega ({{\mathbf{h}}_T})){\mathbf{d}}\omega &= \int q (\omega )\log p({\mathbf{y}}|{\mathbf{f}}_{\mathbf{y}}^\omega ({\mathbf{f}}_{\mathbf{h}}^\omega ({{\mathbf{x}}_T},{{\mathbf{h}}_{T - 1}}))){\mathbf{d}}\omega \hfill \\
&= \int q (\omega )\log p({\mathbf{y}}|{\mathbf{f}}_{\mathbf{y}}^\omega ({\mathbf{f}}_{\mathbf{h}}^\omega ({{\mathbf{x}}_T},{\mathbf{f}}_{\mathbf{h}}^\omega (...{\mathbf{f}}_{\mathbf{h}}^\omega ({{\mathbf{x}}_1},{{\mathbf{h}}_0})...)))){\mathbf{d}}\omega \hfill \\
\end{aligned} \end{equation} \]
Apply Monte Carlo integration with one singe sample:
\[
\approx \log p({\mathbf{y}}|{\mathbf{f}}_{\mathbf{y}}^{\hat \omega }({\mathbf{f}}_{\mathbf{h}}^{\hat \omega }({{\mathbf{x}}_T},{\mathbf{f}}_{\mathbf{h}}^{\hat \omega }(...{\mathbf{f}}_{\mathbf{h}}^{\hat \omega }({{\mathbf{x}}_1},{{\mathbf{h}}_0})...)))),{\text{ }}\hat \omega \sim q(\omega )
\]
Here we have only one sample, so \(q({\hat \omega})=1\).
Plug this estimator into eq. (2), our minimization object becomes:
\[
\begin{array}{*{20}{l}}
\mathcal{L}&{ \approx - \sum\limits_{i = 1}^N {\log } p({{\mathbf{y}}_i}|{\mathbf{f}}_{\mathbf{y}}^{{{\hat \omega }_i}}({\mathbf{f}}_{\mathbf{h}}^{{{\hat \omega }_i}}({{\mathbf{x}}_{i,T}},{\mathbf{f}}_{\mathbf{h}}^{{{\hat \omega }_i}}(...{\mathbf{f}}_{\mathbf{h}}^{{{\hat \omega }_i}}({{\mathbf{x}}_{i,1}},{{\mathbf{h}}_0})...)))) + {\text{KL}}(q(\omega )||p(\omega )).}
\end{array}
\]
The approximating distribution is defined as factorization over rows of weight matrices in model \(\omega \), thus factorized over time steps. For every row \(\mathbf{w}_k\) (corresponding to a time step \(k\)), the approximating distribution is:
\[
q({{\mathbf{w}}_k}){\text{ }} = p\mathbb{N}({{\mathbf{w}}_k};{\mathbf{0}},{\sigma ^2}I) + (1 - p)\mathbb{N}({{\mathbf{w}}_k};{\text{ }}{{\mathbf{m}}_k},{\sigma ^2}I)
\]
where \(\mathbf{m}_k\) is the variational parameter, \(p\) is the dropout probability, \(\sigma^2\) is a small variance.
We have \(q(w)\) for loss function, in other words, we have solved training. What about test time?
For prediction, we can use eq. (1) to average predictions over several samples:
\[
\begin{array}{*{20}{l}}
{p({{\mathbf{y}}^*}|{{\mathbf{x}}^*},{\mathbf{X}},{\mathbf{Y}})}&{ \approx \int p ({{\mathbf{y}}^*}|{{\mathbf{x}}^*},\omega )q(\omega ){\text{d}}\omega \approx \frac{1}{K}\sum\limits_{k = 1}^K p ({{\mathbf{y}}^*}|{{\mathbf{x}}^*},{{\widehat \omega }_k})}
\end{array}
\]
with \({\widehat \omega _k}\sim q(\omega )\).
Implementation in RNNs
The specific practice is to perform dropout in RNNs with the same network units dropped at each time step. This includes recurrent connections too.
For example, LSTM is defined as four gates: input, forget, output, and input modulation:
\[
\begin{equation}
\begin{array}{*{20}{l}}
{\underline {\mathbf{i}} }&{ = {\text{sigm}}({{\mathbf{h}}_{t - 1}}{{\mathbf{U}}_i} + {{\mathbf{x}}_t}{{\mathbf{W}}_i})}&{}&{\underline {\mathbf{f}} = {\text{sigm}}({{\mathbf{h}}_{t - 1}}{{\mathbf{U}}_f} + {{\mathbf{x}}_t}{{\mathbf{W}}_f})} \\
{\underline {\mathbf{o}} }&{ = {\text{sigm}}({{\mathbf{h}}_{t - 1}}{{\mathbf{U}}_o} + {{\mathbf{x}}_t}{{\mathbf{W}}_o})}&{}&{\underline {\mathbf{g}} = {\text{tanh}}({{\mathbf{h}}_{t - 1}}{{\mathbf{U}}_g} + {{\mathbf{x}}_t}{{\mathbf{W}}_g})} \\
{{{\mathbf{c}}_t}}&{ = \underline {\mathbf{f}} \circ {{\mathbf{c}}_{t - 1}} + \underline {\mathbf{i}} \circ \underline {\mathbf{g}} }&{}&{{{\mathbf{h}}_t} = \underline {\mathbf{o}} \circ {\text{tanh}}({{\mathbf{c}}_t})}
\end{array}
\end{equation}
\]
with \(\omega = \{ {{\mathbf{W}}_i},{{\mathbf{U}}_i},{{\mathbf{W}}_f},{{\mathbf{U}}_f},{{\mathbf{W}}_o},{{\mathbf{U}}_o},{{\mathbf{W}}_g},{{\mathbf{U}}_g}\} \) as weight matrices and \(\circ\) as the element-wise product. This model has an alternative form of equations:
\[\begin{equation} \begin{aligned}
\left( {\begin{array}{*{20}{c}}
{\underline {\mathbf{i}} } \\
{\underline {\mathbf{f}} } \\
{\underline {\mathbf{o}} } \\
{\underline {\mathbf{g}} }
\end{array}} \right) &= \left( {\begin{array}{*{20}{c}}
{{\text{sigm}}} \\
{{\text{sigm}}} \\
{{\text{sigm}}} \\
{{\text{tanh}}}
\end{array}} \right)(\left( {\begin{array}{*{20}{c}}
{{{\mathbf{x}}_t}} \\
{{{\mathbf{h}}_{t - 1}}}
\end{array}} \right) \cdot {\mathbf{W}}) \hfill \\
\hfill \\
\end{aligned} \end{equation} \]
with \(\omega={\mathbf{W} }\), where \({\mathbf{W}} \in {\mathbb{R}^{2K \times 4K}},\;{\kern 1pt} {{\mathbf{x}}_t} \in {\mathbb{R}^K} \). Thus,
\[\begin{equation} \begin{aligned}
{\mathbf{W}} &= \left( {\begin{array}{*{20}{c}}
{\begin{array}{*{20}{c}}
{{{\mathbf{W}}_{{\mathbf{\underset{\raise0.3em\hbox{$\smash{\scriptscriptstyle-}$}}{i} }}}}}&{{{\mathbf{W}}_{{\mathbf{\underset{\raise0.3em\hbox{$\smash{\scriptscriptstyle-}$}}{f} }}}}}&{{{\mathbf{W}}_{{\mathbf{\underset{\raise0.3em\hbox{$\smash{\scriptscriptstyle-}$}}{o} }}}}}&{{{\mathbf{W}}_{{\mathbf{\underset{\raise0.3em\hbox{$\smash{\scriptscriptstyle-}$}}{g} }}}}}
\end{array}} \\
{\begin{array}{*{20}{c}}
{{{\mathbf{W}}_{{\mathbf{\underset{\raise0.3em\hbox{$\smash{\scriptscriptstyle-}$}}{i} }}}}}&{{{\mathbf{W}}_{{\mathbf{\underset{\raise0.3em\hbox{$\smash{\scriptscriptstyle-}$}}{f} }}}}}&{{{\mathbf{W}}_{{\mathbf{\underset{\raise0.3em\hbox{$\smash{\scriptscriptstyle-}$}}{o} }}}}}&{{{\mathbf{W}}_{{\mathbf{\underset{\raise0.3em\hbox{$\smash{\scriptscriptstyle-}$}}{g} }}}}}
\end{array}}
\end{array}} \right) \hfill \\
\hfill \\
\end{aligned} \end{equation} \]
In eq. (3), different rows can be dropped, because we have \(4\) separate \(\mathbf{W}\). But in eq. (4), all matrices are tied together. So, only one same dropout mask is allowed. This results in a faster forward-pass, at the sacrifice of slightly diminished results.
More concretely, the later parametrization can be written as
\[\begin{equation} \begin{aligned}
\left( {\begin{array}{*{20}{c}}
{\underline {\mathbf{i}} } \\
{\underline {\mathbf{f}} } \\
{\underline {\mathbf{o}} } \\
{\underline {\mathbf{g}} }
\end{array}} \right) &= \left( {\begin{array}{*{20}{c}}
{{\text{sigm}}} \\
{{\text{sigm}}} \\
{{\text{sigm}}} \\
{{\text{tanh}}}
\end{array}} \right)(\left( {\begin{array}{*{20}{c}}
{{{\mathbf{x}}_t}\circ {{\mathbf{z}}_{\mathbf{x}}}} \\
{{{\mathbf{h}}_{t - 1}}\circ {{\mathbf{z}}_{\mathbf{h}}}}
\end{array}} \right) \cdot {\mathbf{W}}) \hfill \\
\hfill \\
\end{aligned} \end{equation} \]
where \({{{\mathbf{z}}_{\mathbf{x}}}}\) and \({{{\mathbf{z}}_{\mathbf{h}}}}\) are random masks at all time steps.
Word Embeddings Dropout
It' common to see dropout in embedding layer, which randomly zeros some rows in word vector. But in this paper, the author suggested to zero entire word vector based on its type. For example, if word "the" is dropped, then sentence "the dog and the cat" becomes "0 dog and 0 cat". But there will never be a sentence like "0 dog and the cat".
References
- Gal, Y., & Ghahramani, Z. (2016). A theoretically grounded application of dropout in recurrent neural networks. In Advances in neural information processing systems (pp. 1019-1027).



